The most commonly used names for enzymes have "―ase" attached to the substrate of the Glucosidase reaction. For example, glucosidase is the enzyme that acts on glucose. Alternatively, enzyme names can contain a description of the performed action, as seen in lactate dehydrogenase, which catalyzes the conversion of lactate to pyruvate.
The systematic name for pepsin is endopeptidase, which reflects its function in breaking down proteins into smaller peptides in the stomach.
Some enzymes retain their original trivial names. The systematic name for pepsin. The systematic name for pepsin is peptidyl peptide hydrolase. This name reflects its function as an enzyme that breaks down peptide bonds in proteins during digestion.
The International Union of Biochemistry and Molecular Biology (IUBMB) has developed a nomenclature system that classifies enzymes into six main classes based on the type of reaction they catalyze.
| № | Class Name | Characterization of Reactions | Class Representatives |
|---|
| 1 | Oxidoreductases | Catalyze oxidation-reduction reactions (transfer of electrons) | Lactate dehydrogenase |
| 2 | Transferases | Transfer functional groups (e.g., methyl or phosphate groups) | Alanine transaminase |
| 3 | Hydrolases | Catalyze the hydrolysis of various bonds | Pepsin, lipase |
| 4 | Lyases | Catalyze the addition or removal of groups to form double bonds | Pyruvate decarboxylase |
| 5 | Isomerases | Catalyze isomerization changes within a single molecule | Phosphoglucose isomerase |
| 6 | Ligases | Catalyze the joining of two molecules with the use of ATP | DNA ligase |
In enzyme kinetics, the reaction rate is measured and the factors influencing this rate are investigated. Studying the kinetics of an enzyme in this way can reveal the catalytic mechanism of the enzyme, its role in metabolism, how its activity is controlled, and how a drug or agonist can inhibit the enzyme.
In enzyme kinetics, the reaction formula typically follows the Michaelis-Menten equation, which is expressed as:
Explanation of Each Part of the Formula:
- v: This represents the initial reaction velocity or rate of the enzyme-catalyzed reaction. It indicates how fast the substrate is converted into product at a given substrate concentration.
- Vmax : This is the maximum reaction velocity that can be achieved by the system at saturating substrate concentrations. It reflects the maximum rate at which the enzyme can convert substrate into product when all active sites are occupied.
- [S]: This denotes the substrate concentration. It is the amount of substrate available for the enzyme to act upon. The reaction rate depends on this concentration, especially at lower levels.
- Km: This is the Michaelis constant, a key parameter that provides insight into the enzyme's affinity for its substrate. It is defined as the substrate concentration at which the reaction velocity is half of Vmax. A low Km value indicates high affinity, meaning the enzyme can achieve half-maximal velocity at a low substrate concentration.
A short preview of Michaelis-Menten equation.
The Michaelis-Menten equation is fundamental in enzyme kinetics as it describes how the rate of an enzyme-catalyzed reaction depends on substrate concentration. By analyzing this equation, researchers can gain insights into the enzyme's catalytic mechanism, its role in metabolic pathways, and how various factors, including inhibitors or activators, can influence enzyme activity.
The reaction mechanism for enzyme-catalyzed reactions can be represented by the following equation:
E+S⇌ES→E+P
Explanation of Each Part of the Reaction:
E: This represents the enzyme. Enzymes are biological catalysts that speed up chemical reactions without being consumed in the process.
S: This denotes the substrate, which is the molecule upon which the enzyme acts. The substrate binds to the enzyme to form a complex.
ES: This is the enzyme-substrate complex. It forms when the substrate binds to the active site of the enzyme. This complex is a crucial intermediate in the reaction, as it is where the actual conversion of substrate to product occurs.
P: This represents the product of the reaction. After the enzyme catalyzes the conversion of the substrate, the product is released, and the enzyme is regenerated in its original form.
Reaction Steps:
Formation of the Enzyme-Substrate Complex: The enzyme E binds to the substrate S to form the enzyme-substrate complex ES. This step is often reversible, meaning that the substrate can dissociate from the enzyme without being converted into product.
Conversion to Product: The enzyme-substrate complex ES undergoes a transformation to produce the product P. This step is typically irreversible under physiological conditions, as it leads to the release of the product and the regeneration of the free enzyme E.
Graph of the Dependence of the Reaction Rate on:
Temperature: The graph typically shows that as temperature increases, the reaction rate increases up to an optimal temperature, after which the rate declines due to enzyme denaturation.
pH: The graph illustrates that each enzyme has an optimal pH range. The reaction rate increases within this range and decreases outside of it, often showing a bell-shaped curve.
Substrate Concentration: The graph demonstrates that initially, as substrate concentration increases, the reaction rate increases. However, it eventually levels off as the enzyme becomes saturated, reaching Vmax.
A. Enzyme Inhibition
Enzyme inhibition is the process by which a molecule (inhibitor) decreases the activity of an enzyme, preventing it from catalyzing a reaction effectively.
Inhibition Types of an typical enzyme:
Competitive Inhibition: The inhibitor competes with the substrate for binding to the active site of the enzyme.
Non-competitive Inhibition: The inhibitor binds to an enzyme at a site other than the active site, reducing the enzyme's activity regardless of substrate concentration.
Uncompetitive Inhibition: The inhibitor binds only to the enzyme-substrate complex, preventing the conversion of substrate to product.
B. Enzyme Specificity
Enzyme specificity refers to the ability of an enzyme to select and catalyze a specific substrate or group of substrates, ensuring that the correct biochemical reactions occur in the cell.
C. Types of Specificity:
Absolute Specificity: The enzyme catalyzes only one specific substrate.
Group Specificity: The enzyme acts on a group of substrates that have similar chemical structures.
Linkage Specificity: The enzyme catalyzes reactions involving specific types of chemical bonds, regardless of the substrate's overall structure.
D. Structure of Enzymes:
- Holoenzyme: The complete, active form of an enzyme that includes the apoenzyme and its cofactor(s).
- Apoenzyme: The protein portion of an enzyme, which is inactive without its cofactor.
- Cofactor: A non-protein chemical compound that is required for the enzyme's activity, which can be a metal ion or a small organic molecule.
- Coenzyme: A specific type of cofactor that is organic and often derived from vitamins, assisting in the enzyme's catalytic activity.


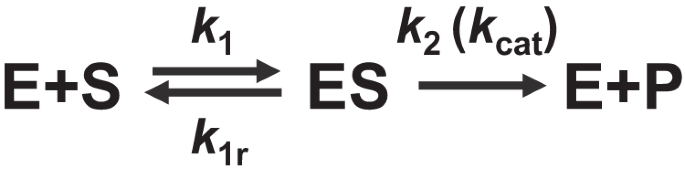
Post a Comment